Researchers from Griffith University’s Institute for Glycomics in Australia, together with the Nationwide Children’s Hospital in Ohio, have uncovered evidence to help vaccine developers prevent middle ear infections, or otitis media.
According to an article published in the July 28, 2015 edition of Nature Communications, the researchers found that the bacterial pathogen Haemophilus influenzae, which causes illnesses such as pneumonia, chronic obstructive pulmonary disease, and otitis media, contains a system that randomly changes gene expression causing the bacteria to switch between two different cell types. The authors said that this “moving target” is what has made vaccine development difficult in the past.
A principal research leader at the Institute for Glycomics, Michael Jennings, PhD, said that middle ear infections, or otitis media, are the most frequent reason children see a doctor or undergo pediatric surgical procedures.
“The treatment of otitis media is currently with antibiotics and the insertion of ear tubes (tympanostomy),” said Jennings. “If there was a vaccine developed, this would dramatically reduce the amount of antibiotics prescribed. Through this research we have been able to understand the lifestyle of the bug and its adaptation to us as hosts, and therefore we now have a better idea of which surface proteins are good targets for vaccine development.”
Jennings added that their findings are a very important step in developing a vaccine and will save developers a considerable amount of time and money.

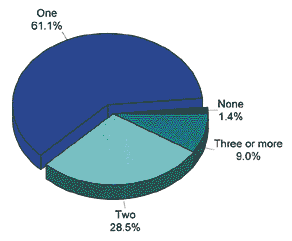
This figure shows analysis of NTHi strain collections to ascertain the proportion of isolates containing phase variable modA alleles.
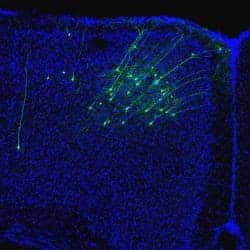
In the study, the researchers sought to assess the role that candidate non-typeable Haemophilus influenzae (NTHi)–a significant bacterial pathogen commonly associated with otitis media–may play in gene expression, immunoevasion, and in the pathogenesis of NTHi during experimental otitis media (OM).
The researchers demonstrated that phase variation of all five of the modA alleles for NTHi control gene expression differences in all the strains studied. These expression differences include outer-membrane proteins and key vaccine candidates. According to the article, the study showed that biphasic switching of specific modA alleles also influences fitness in opsonophagocytic killing assays and in susceptibility to antibiotics.
Jennings said the next step is to see if this discovery might relate to other illnesses caused by Haemophilus influenzae, such as chronic obstructive pulmonary disease. Jennings worked with Lauren Bakaletz, PhD, and her team at The Research Institute at Ohio’s Nationwide Children’s Hospital, along with a team from Griffith’s Institute for Glycomics that included John Atack, PhD.
Source: The Institute for Glycomics, Griffith University, Australia
Image credits: Griffith University, Nature Communications